Docket #: S17-020
MAGESTIC - A High Efficiency, Massively Parallel Production of Genetically Engineered Clones for Functional Genomics and Synthetic Biology
Researchers in the Stanford Genome Technology Center have developed a robust, high-throughput, high-efficiency functional genomics platform to generate precisely edited genome variant libraries and then readily isolate and identify thousands of individual strains en masse from a mutant pool. This technology, called MAGESTIC (multiplexed accurate genome editing with short, trackable, integrated cellular barcodes), combines RNA-guided nuclease genome editing (e.g., CRISPR) with stable, integrated barcoding to enable marker-free variant tracking, quantitative read out of phenotypic effects of DNA variants, and strain isolation via recombinase directed indexing, a.k.a. REDI. In particular, by providing access to individual sequence-validated strains, the variants within the library can be characterized with a wide range of assays, including microscopy localization, metabolomics and enzyme function. Additional features of this system include 99% gene editing efficiency and the ability to perform complete saturation mutagenesis of protein-coding regions.
Overall, MAGESTIC enables tens of thousands of specific genetic variants across the genome to be created in a manner that is compatible with robust phenotyping across hundreds of conditions.
This genome engineering approach could be used in basic research to significantly advance our understanding of the genotype-environment-phenotype relationship. In addition, MAGESTIC could be used in synthetic biology applications to engineer enzymes or organisms for optimized industrial, agricultural or medicinal applications.
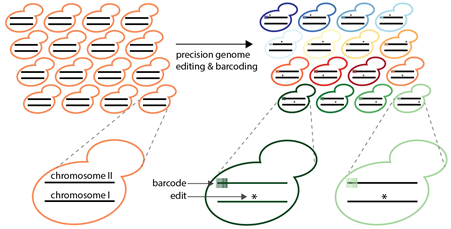
Schematic of MAGESTIC (Multiplex precision genome editing with short, trackable, genomic barcodes) – Combining barcoding with high throughput, programmable genome editing enables isolation of individual, sequence validated strains following transformation with a complex library.
Stage of Research
The inventors have demonstrated the utility of this platform in yeast with saturation editing of an essential gene followed by fine-scale dissection to identify amino acids critical for chemical inhibition of a lipid signaling protein. Furthermore, they showed genome-wide capacity by introducing thousands of naturally occurring genetic variants across the entire genome. Their continued work is focused on scaling the technology up so that they can introduce hundreds of thousands of variants throughout the genome.
Applications
- Synthetic biology - rapidly generate libraries of variant strains with end user applications such as protein and metabolic engineering to generate desirable traits for industrial, agricultural and medicinal applications
- Research:
- Perform high-throughput functional genomics studies, particularly amenable to microscopy, metabolomics and enzymatic assay analysis
- Dissect genetic basis of quantitative traits
Advantages
- High throughput with isolation of individual strains - engineers precise variants at the genome scale
- lowers cost of sequence-validated variant library production
- parses thousands of individual variants en masse from a mutant pool using barcodes with recombinase directed indexing (REDI)
- identifies precise genetic modifications
- enables wider range of techniques to characterize variants, such as high content microscopy, enzymatic assays and metabolomics
- Highly efficient editing:
- editing efficiency >99% (>5-fold increase with active donor recruitment using the LexA-Fkh1p system)
- active donor DNA-directed recruitment increases recombination efficiency and favors precision editing
- can create single nucleotide variants not linked to PAM mutations
- internal marker removes background due to oligonucleotide synthesis errors
- edits codons outside of guide RNA to enable complete saturation mutagenesis of protein-coding regions
- Quantitative read-out - stable, integrated barcodes (instead of plasmid barcodes) enable marker-free variant tracking and one-to-one correspondence of barcode counts with abundance for quantitative read-out of phenotypic effects
- Flexible platform - basic MAGESTIC system and reagents can be adapted for use with any organism or cell lines, such as mammalian cells or bacteria
Publications
- Kevin R. Roy, Justin D. Smith, Sibylle C. Vonesch, Gen Lin, Chelsea Szu Tu, Alex R. Lederer, Angela Chu, Sundari Suresh, Michelle Nguyen, Joe Horecka, Ashutosh Tripathi, Wallace T. Burnett, Maddison A. Morgan, Julia Schulz, Kevin M. Orsley, Wu Wei, Raeka S. Aiyar, Ronald W. Davis, Vytas A. Bankaitis, James E. Haber, Marc L. Salit, Robert P. St. Onge, Lars M. Steinmetz, "Multiplexed precision genome editing with trackable genomic barcodes in yeast," Nature Biotechnology, May 7, 2018.
Related Publication for REDI Technology
Smith, J. D., Schlecht, U., Xu, W., Suresh, S., Horecka, J., Proctor, M. J., ... & Roy, K. (2017). A method for high-throughput production of sequence-verified DNA libraries and strain collections. Molecular systems biology, 13(2), 913.
Related Links
Patents
- Published Application: WO2019055878
- Published Application: 20200270632
Similar Technologies
-
High-throughput precision genome editing S16-025High-throughput precision genome editing
-
A multiplexed RNA regulation platform for primary immune cell engineering S22-235A multiplexed RNA regulation platform for primary immune cell engineering
-
Large Scale Genomic Editing and Tracking using CRISPR-based Single-Cell Barcoding S19-349Large Scale Genomic Editing and Tracking using CRISPR-based Single-Cell Barcoding