Docket #: S14-476
Modular system for programming protein form and function
Researchers in Prof. Christina Smolke's laboratory have engineered a conditional protein expression platform that enables complex regulatory programming in higher organisms through control of alternative splicing. This “Alternative Splicing Device” (ASD) system uses RNA control elements that can be switched on and off with specific ligands. These switches combined with modular exons code for different functional domains. Together, the ASD can be used to regulate the expression of an assortment of different protein isoforms from a single messenger sequence. Because the RNA switches can detect a variety of intracellular and extracellular signals (such as small molecules, proteins or nucleic acids), they can dynamically program protein function for the regulated production of precise protein forms, activity, or locations. The ASD could be used in a variety of research or therapeutics applications such as transcriptional control, signal transduction, and cellular imaging.
Stage of Research
Researchers are using this platform with improved splicing regulators for controlling mutually exclusive alternative splicing.
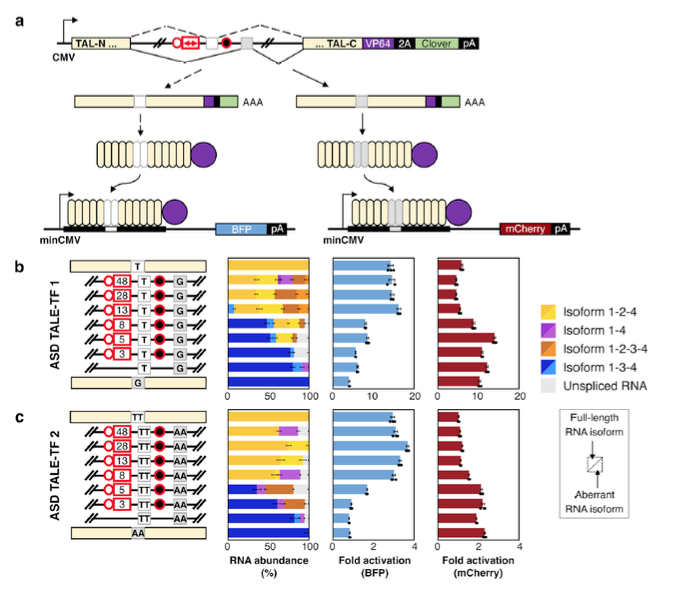
Spliced transcription factors generate mutually exclusive isoforms for controlled gene activation from two distinct promoters. a Schematic illustrating the activation of two fluorescent proteins from a single MEAS event, where isoform 1-2-4 activates BFP reporter expression and isoform 1-3-4 activates mCherry reporter expression. Devices contain mutated BP elements for eons 2 and 3 (circles with red outlines) and modified exon 2 PPT elements (white rectangle with red outline representing PPT sequence lengths in nucleotides). b ASD TALE-TF1 devices built from a modified intron framework that encode RVD NG (target site T) in exon 2 and RVD NN (target site G) in exon 3. c ASD TALE-TF 2 devices built from a modified intron framework that encode RVDs NG NG (target site TT) in exon 2 and RVDs NI NI (target site AA) in exon 3. Relative RNA isoform abundances were characterized using long-read sequencing. Error bars in the RNA isoform data represent ±1 standard deviation from biological triplicates. Blue represents RNA isoform 1-3-4 and its derivatives; yellow represents RNA isoform 1-2-4 and its derivatives; orange represents RNA isoform 1-2-3-4 and its derivatives; purple represents RNA isoform 1-4 and its derivatives; and gray represents unspliced transcript. The fold activation was determined via flow cytometry analysis of BFP and mCherry fluorescence in transfected HEK-293T cells, and calculated as the ratio of the median BFP or mCherry fluorescence intensity of cells co-transfected with and without the specified ASD TALE-TF. Fold activations from biological triplicates were averaged and reported within an error range of ±1 standard deviation.
Applications
- Protein expression system - modular system for programmable protein expression to build synthetic pathways and control production of protein variants, with ultimate applications for:
- transcriptional control - modular transcription factors (such as TALENs) can be programmed to control expression of endogenous genes for cell therapy, targeted cancer treatments, or basic researh
- signal transduction - enzymes can be engineered to trigger, alter, or amplify cell signaling
- subcellular localization - specific domains can be spliced in or out of protein to direct it to particular sites within a cell
- cellular imaging - with conditional splicing of engineered fluorescent proteins
Advantages
- Controls form, function and expression - the platform supports control beyond traditional gene expression and could include spatial organization or protein activity
- Modular and extensible:
- basic framework of intronic RNA switches can be combined with protein domains of choice
- any gene can be split into artificial exons to form functional domains that can be alternatively spliced into the protein end product
- precise splicing to control how exons are combined enables numerous proteins to be encoded
- Independent regulation - regulatory information is placed in non-protein coding regions so they do not disturb the encoded protein
- Multiple switches for programmable expression - can be responsive to intracellular or extracellular signals (such as small molecules, proteins, DNA or RNA oligonucleotides) which expand the inputs the device can sense and process to produce a response
Publications
- M. Mathur, C.M. Kim, S.A. Munro, S.S. Rudina, E.M. Sawyer and C.D. Smolke Programmable mutually exclusive alternative splicing for generating RNA and protein diversity Nature Communications (2019)10:2673.
Related Links
Patents
- Issued: 10,053,697 (USA)
Similar Technologies
-
DICE: dual-integrase cassette exchange S13-057DICE: dual-integrase cassette exchange
-
Gene sequences for the biosynthesis of podophyllotoxin derivatives S15-017Gene sequences for the biosynthesis of podophyllotoxin derivatives
-
A Method to Regulate Protein Function in Living Cells Using Small Molecules S06-024A Method to Regulate Protein Function in Living Cells Using Small Molecules