Docket #: S16-369
CLOuD9- a method for CRISPR-mediated chromosomal looping
Researchers at Stanford and their collaborators have developed a targeted, reversible method to manipulate chromatin structure. Chromatin folding within the nucleus and, further, the chromosomal contacts created by the 3D organization of chromatin are critical to gene expression regulation. As such, it would be beneficial to have a comprehensive understanding of the functional organization and formation of chromatin loops. However, this is lacking as there are no tools to help achieve such an understanding. To overcome this limitation the inventors have developed CLOuD9 (Chromosomal Loop Reorganization using CRISPR-dCas9) to selectively and reversibly establish chromatin loops. CLOuD9 uses nuclease deficient Cas9 variants fused to reversible, chemically-induced dimerization proteins and standard guide RNAs (gRNAs) to direct the dCas9 components to the targeted locus. Addition of the chemical inducer forces juxtaposition of the targeted genomic loci to create a loop. This technology provides a broadly applicable method to modify chromatin loops and selectively modulate gene expression.
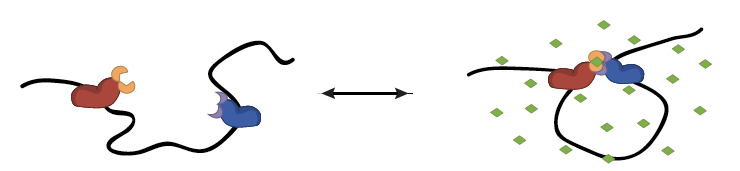
Schematic of CLOuD9 induced looping. Addition of abscisic acid (ABA, green) brings two complementary CLOuD9 constructs (red and blue) into proximity, remodeling the chromatin structure. Removal of ABA restores the endogenous chromatin conformation.
Stage of research
The inventors have used the method to show that modification of the chromatin loop structure alters gene expression.
Applications
- Research tool- study relationship between chromatin structure and gene expression
Advantages
- Solves an unmet need- provides a powerful tool for precise chromatin loop modification
- Method is:
- Targeted
- Selective
- Reversible
- Easy-to-use
- Does not require manipulation of the underlying DNA
- Can be applied anywhere CRISPR gRNAs can target
Publications
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, Hoffman AR, Huang CC, Pitteri SJ, Wang KC. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017 Jul 13;8:15993.
Patents
- Published Application: 20180355344
- Issued: 11,293,018 (USA)
Similar Technologies
-
Methods for Generating Pooled RNA Oligos for Multiplexed Chromosome Targeting, Imaging, and Perturbation S24-492Methods for Generating Pooled RNA Oligos for Multiplexed Chromosome Targeting, Imaging, and Perturbation
-
High-throughput precision genome editing S16-025High-throughput precision genome editing
-
Super Recombinator (SuRe), a CRISPR/Cas9-based platform for the rapid incorporation of multiple transgenes at the same genetic locus S20-282Super Recombinator (SuRe), a CRISPR/Cas9-based platform for the rapid incorporation of multiple transgenes at the same genetic locus