Docket #: S18-500
Methods for improved prediction of cellular differentiation states from single cell genomic data
Researchers at Stanford have developed a computational tool that enables the discovery of regenerative cells across all tissue types and novel targets in cancer. The tool, which uses single-cell RNA sequencing (scRNA-seq) data as input, is called CytoTRACE and is based on the premise that less differentiated cells express a larger number of genes.
CytoTRACE has been validated across 42 scRNA-seq datasets and outcompeted ~19,000 other features in predicting the differentiation status of single cells. The platform can identify markers to isolate regenerative cells in normal human tissues and target tumor-initiating cells that drive recurrence in cancer. CytoTRACE can be applied independently of tissue, platform, and species, with no prior knowledge required.
The inventors have validated their discovery pipeline in data from human breast cancer patients. CytoTRACE identified a new target, GULP1, and demonstrated that the knockdown of this gene abrogates the growth of human breast cancers xenografted in mice.
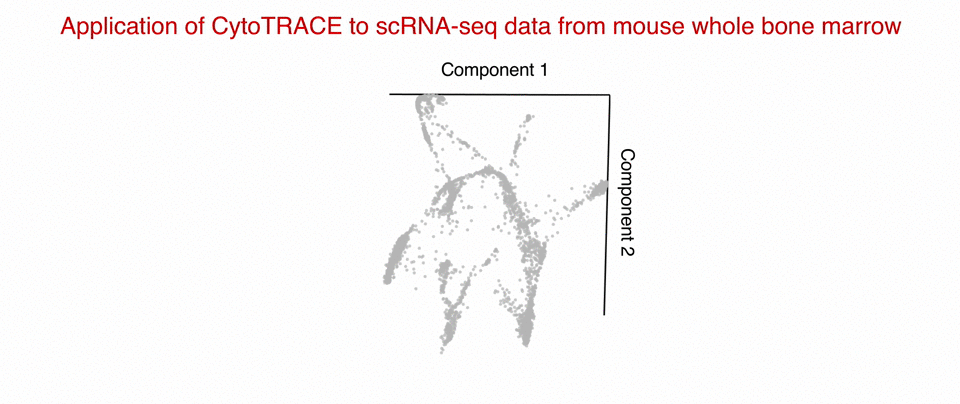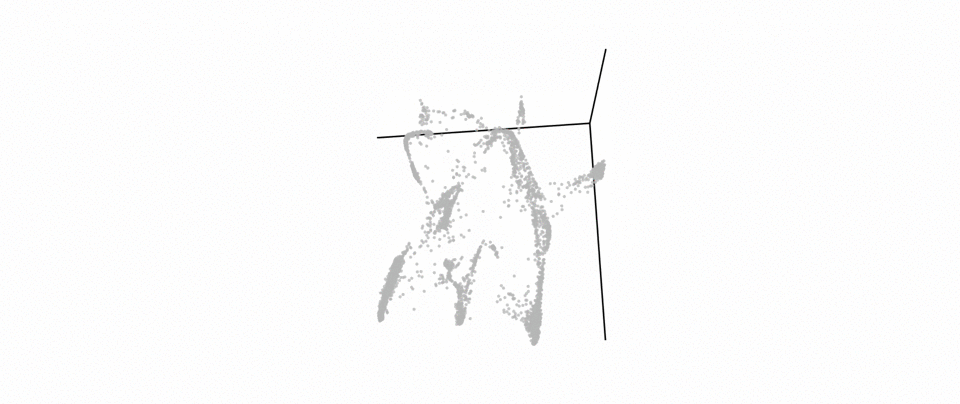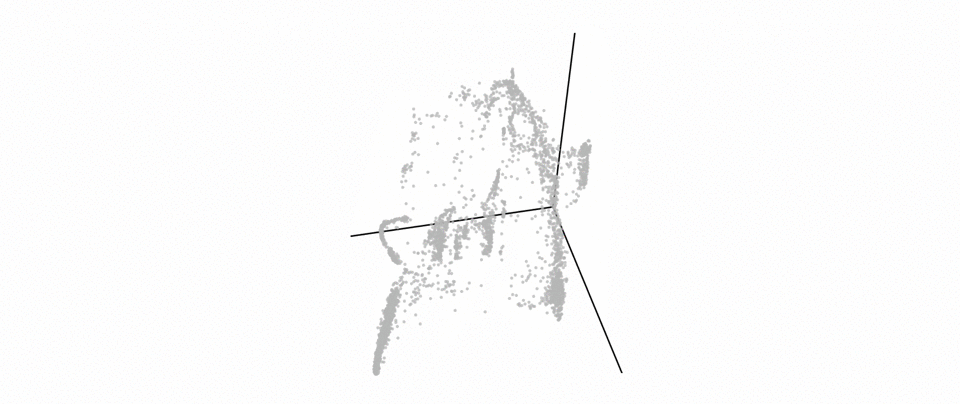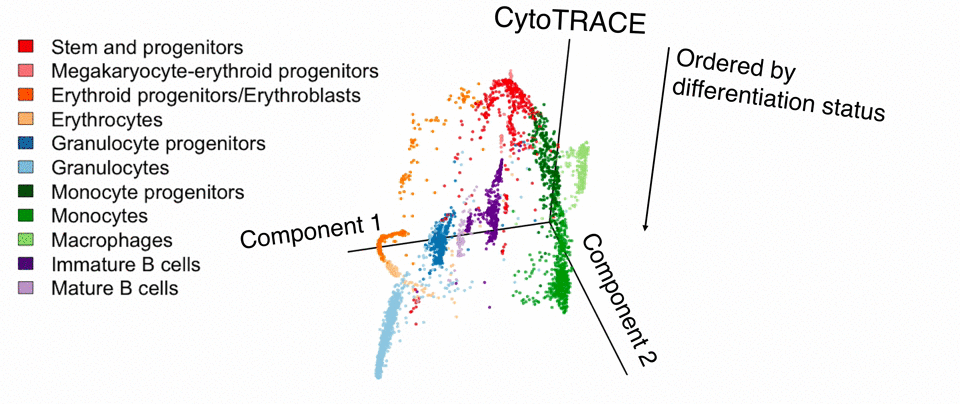
Stage of Development: In vivo - abrogates the growth of human breast cancers xenografted in mice
Applications
- Target discovery in oncology and regenerative medicine
- Identification of tumor-initiating cells
- Identification of novel stem cell and tumor cell populations
- Biomarker identification in oncology and cellular regeneration
- Characterization of cellular hierarchies in complex cell populations
- Assessment of chromatin accessibility on a single-cell level
Advantages
- Applicable across all cell types, tissues, and species
- Can be applied independently of sequencing protocol
- No need for a 'reference' point or prior knowledge of differentiation status
- Outperforms many leading computational models
Publications
- Gulati G. et al. Single-cell transcriptional diversity is a hallmark of developmental potential Science 2020.
Related Links
Patents
- Published Application: 20200370112
- Issued: 12,460,260 (USA)
Similar Technologies
-
CytoTrace2: Methods and Systems for Determining Phenotypic States from Genomic Data with Interpretable AI S24-057CytoTrace2: Methods and Systems for Determining Phenotypic States from Genomic Data with Interpretable AI
-
High-throughput screening of 3D genome architecture using Plate-C S25-094High-throughput screening of 3D genome architecture using Plate-C
-
A Method to Regulate Protein Function in Living Cells Using Small Molecules S06-024A Method to Regulate Protein Function in Living Cells Using Small Molecules