Docket #: S22-281
Multiplex Epstein-Barr Virus Genotyping Nucleic Acid Amplification Test for Detection of High-risk Variants in Human Plasma
Researchers at Stanford have developed a nucleic acid amplification test to detect high-risk Epstein-Barr Virus (EBV) BALF2 variants in plasma to aid population-level screening for nasopharyngeal carcinoma (NPC). The team designed and validated a multiplex allele-specific real-time polymerase chain reaction (qPCR) genotyping assay to detect three EBV BALF2 variants. As compared to other current assays, this assay is single-reaction, cost-effective, more accurate, and can also provide important genotyping information. It has been through rigorous analytical/clinical validation.
Commercial application can be in the form of a qPCR kit (analyte-specific reagents or complete kit), which could be purchased by laboratories that offer plasma-based NPC screening.
Stage of Development
Analytically and clinically validated assay
Figures
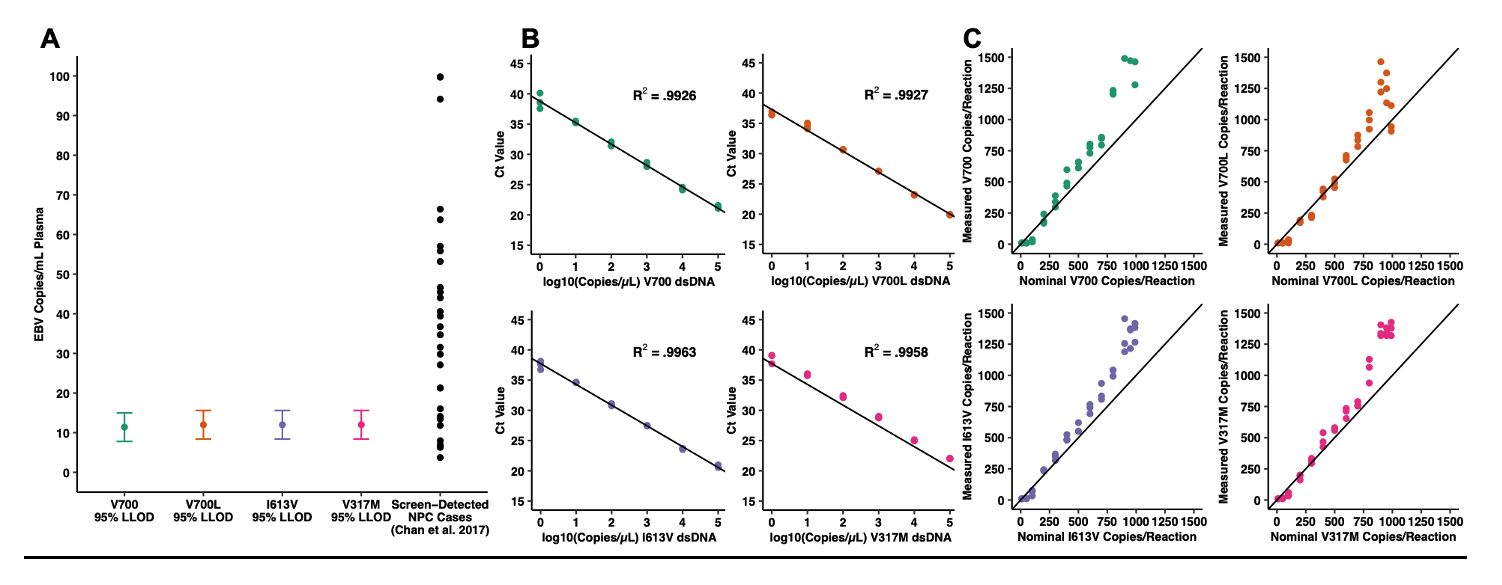
Figure Description A, B, C: Multiplex EBV BALF2 genotyping qPCR design, validation, and association studies with nasopharyngeal carcinoma in endemic and non-endemic populations. A) Analytical sensitivity for each of the four BALF2 qPCR targets. The 95% lower limit of detection with 95% confidence interval is reported for each target in units of EBV copies/mL plasma. In conjunction with the LLODs, the corresponding plasma viral load for 34 screen-detected preclinical NPC cases is presented to indicate likelihood of genotyping success. B) Analytical linearity for each of the four BALF2 qPCR targets, plotting cycle threshold (Ct) against nominal dsDNA control concentration in units of log10 copies/?L template. C) Mixing studies at fixed total template concentration (100 copies/?L template) combining high-risk and low-risk dsDNA controls, demonstrating detection of minor allele fractions as low as 10% for each of the four targets. Measured concentration is plotted against nominal concentration. In the presence of mixed alleles, the assay is approximately linear as allele fraction decreases.
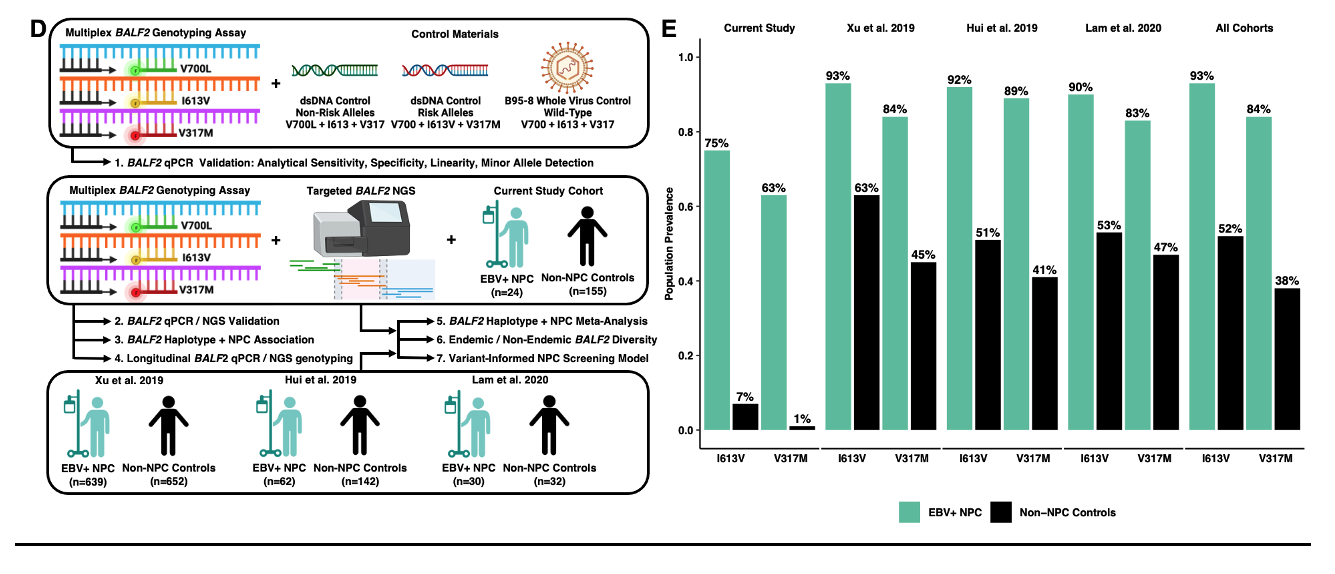
Figure Description D, E: Multiplex EBV BALF2 genotyping qPCR design, validation, and association studies with nasopharyngeal carcinoma in endemic and non-endemic populations. D) Study overview and experimental workflow. First, the multiplex BALF2 genotyping assay was analytically validated using synthetic dsDNA controls and wild-type B95-8 whole virus control. Next, our non-endemic cohort of 24 NPC cases and 155 non-NPC controls contributed to BALF2 qPCR/NGS validation, longitudinal BALF2 genotyping, and BALF2-NPC association. Finally, our non-endemic cohort and three predominantly endemic cohorts contributed to a meta-analysis of 755 EBV+ NPC cases and 981 non-NPC controls. This validated the association between BALF2 haplotypes and NPC in multiple cohorts, further defined regional EBV genomic diversity, and was used to develop a variant-informed screening model. E) Prevalence of I613V and V317M between EBV+ NPC cases and non-NPC controls in the present study and in the three prior EBV GWAS cohorts.
Applications
- Early cancer detection especially in higher-risk East/Southeast Asian populations
- Flexibility for serial or once-lifetime screening triage
- Commercial application can be in the form of a qPCR kit (analyte-specific reagents or complete kit), which could be purchased by laboratories that offer plasma-based nasopharyngeal carcinoma screening.
Advantages
- Earlier cancer detection for better outcomes
- More accurate - reduces false positives which lead to unnecessary procedures
- Cost effective and facilitates both serial or once-lifetime testing
- Single reaction design and flexibility for use on any real-time PCR instrument
- Differentiates EBV BALF2 genotype using three loci and also contains internal control.
- Completed rigorous analytical/clinical validation
Publications
- Miller, J.A., Sahoo, M.K., Yamamoto, F., Huang, C., Wang, H., Zehnder, J.L., Le, Q.T. and Pinsky, B.A. (2022). Multiplex Epstein-Barr virus BALF2 genotyping detects high-risk variants in plasma for population screening of nasopharyngeal carcinoma. Molecular Cancer, 21(1), pp.1-10.
Related Links
Patents
- Published Application: WO2024026336
- Published Application: 20250340958
Similar Technologies
-
Method for Improved Pathogen Detection S18-367Method for Improved Pathogen Detection
-
High-specificity proximity ligation assay to enable detection with low-affinity agents S16-365High-specificity proximity ligation assay to enable detection with low-affinity agents
-
Gene Set for the Diagnosis of Active Pulmonary Tuberculosis S15-057Gene Set for the Diagnosis of Active Pulmonary Tuberculosis