Docket #: S15-409
SNAIL-RCA: A method for multiplexed RNA visualization in single cells
Researchers at Stanford have developed the SNAIL-RCA method for inexpensive and efficient multiplexed detection of single RNA molecules in single cells. SNAIL-RCA (Splint Nucleotide Assisted Intramolecular Ligation followed by Rolling Circle Amplification) provides a simplified proximity ligation technique to enable multiplexed, quantitative, targeted detection of single molecules of RNA in single cells, fresh-frozen tissue and archival FFPE sections. The method may be combined with flow cytometry or mass cytometry to simultaneously analyze large numbers of cells from multiple nucleic acids (up to 20 or more transcripts can be simultaneously analyzed, at a rate of up to ~500 or more cells/second). The RCA products can be detected with a variety of techniques including optical microscopy and SIMS ion beam imaging. This technology will aide biomarker discovery and enable development of molecular diagnostics, including cancer diagnostics, for precision medicine.
Stage of Development
The researchers have demonstrated that SNAIL-RCA enables easily configured, diligently managed, quantitative detection of gene expression levels in single cells.
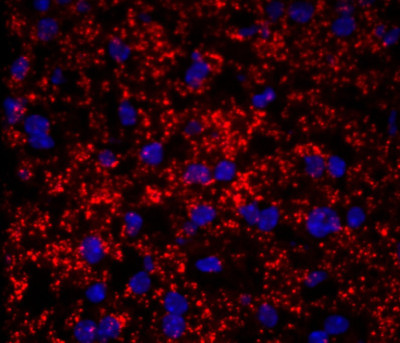
Applications
- Multiplexed and quantitative gene expression profiling in single cells for:
- Molecular diagnostics for precision medicine
- Biomarker discovery
- Research
Advantages
- Allows simultaneous analysis of multiple nucleic acids and proteins in a single cell
- Simple design of SNAIL-RCA:
- Requires only a handful of probe pairs per gene
- Allows for simple configuration of oligonucleotides
- Eliminates need for boilerplate hybridization-ligation sequences
- Reduces the overall reagent cost per gene
- Streamlined proximity ligation protocol
- Efficient detection of small RNAs
- Method is compatible with conventional antibody staining
- Opens the possibility for much higher degree of multiplexing
Related Links
Patents
- Published Application: WO2017147483
- Published Application: 20190055594
- Published Application: 20190093156
- Published Application: 20210238665
- Issued: 11,008,608 (USA)
- Issued: 10,982,271 (USA)
Similar Technologies
-
CytoTrace2: Methods and Systems for Determining Phenotypic States from Genomic Data with Interpretable AI S24-057CytoTrace2: Methods and Systems for Determining Phenotypic States from Genomic Data with Interpretable AI
-
MACHETE – algorithm for automated detection of RNA gene fusions S16-250MACHETE – algorithm for automated detection of RNA gene fusions
-
Low-cost, Comprehensive Methylation Profiling Using Low DNA Inputs for Cancer Diagnostics S23-456Low-cost, Comprehensive Methylation Profiling Using Low DNA Inputs for Cancer Diagnostics