Docket #: S22-109
Direct Coupling of Biolayer Interferometry and Mass Spectrometry for Top-Down Protein Analysis
Researchers at Stanford University have developed an affinity capture technique for top-down protein analysis that directly couples biolayer interferometry (BLI) with high resolution mass spectrometry (HR-MS).
Top-down mass spectrometry (TD-MS) is widely used to provide structural information, such as post-translational modifications and proteoform variation, on intact proteins. While a powerful method for characterizing intact protein structure, the quality of data acquired during TD-MS is highly dependent on the sample preparation. To prepare samples for MS analysis, protein analytes are typically purified from complex biological mixtures using an affinity (or immunoaffinity) capture technique on beads or resin. Yet, conventional affinity capture lacks real-time process monitoring, making optimization of sample preparation in case of weak or absent MS signal challenging. Alternatively, Biolayer Interferometry (BLI), a label-free optical sensing technology, is widely used to study the kinetics and affinity of real-time biomolecular interactions. BLI necessarily monitors the affinity capture of analytes in real-time, but cannot provide any structural information on the captured binding partners. Thus, the combination of BLI and TD-MS can provide both real-time monitoring of protein capture and structural identification. However, development of a seamless coupling method between these two technologies has been hindered by the small sample quantity that can be captured on the BLI microprobe tips.
To address this challenge, this technology directly couples BLI with HR-MS with a method called Microprobe-Capture In-Emitter Elution (MPIE). To implement MPIE, the analyte is first captured on the surface of a microprobe (Figure 1A), the microprobe is directly inserted into an electrospray emitter, and the analyte is eluted by spraying into a mass spectrometer for HR-MS analysis (Figure 1B). In comparison to conventional affinity capture techniques such as bead-based immunoprecipitation, MPIE introduces real-time process monitoring and provides binding characteristics of analytes, offering more information-rich experimental results.
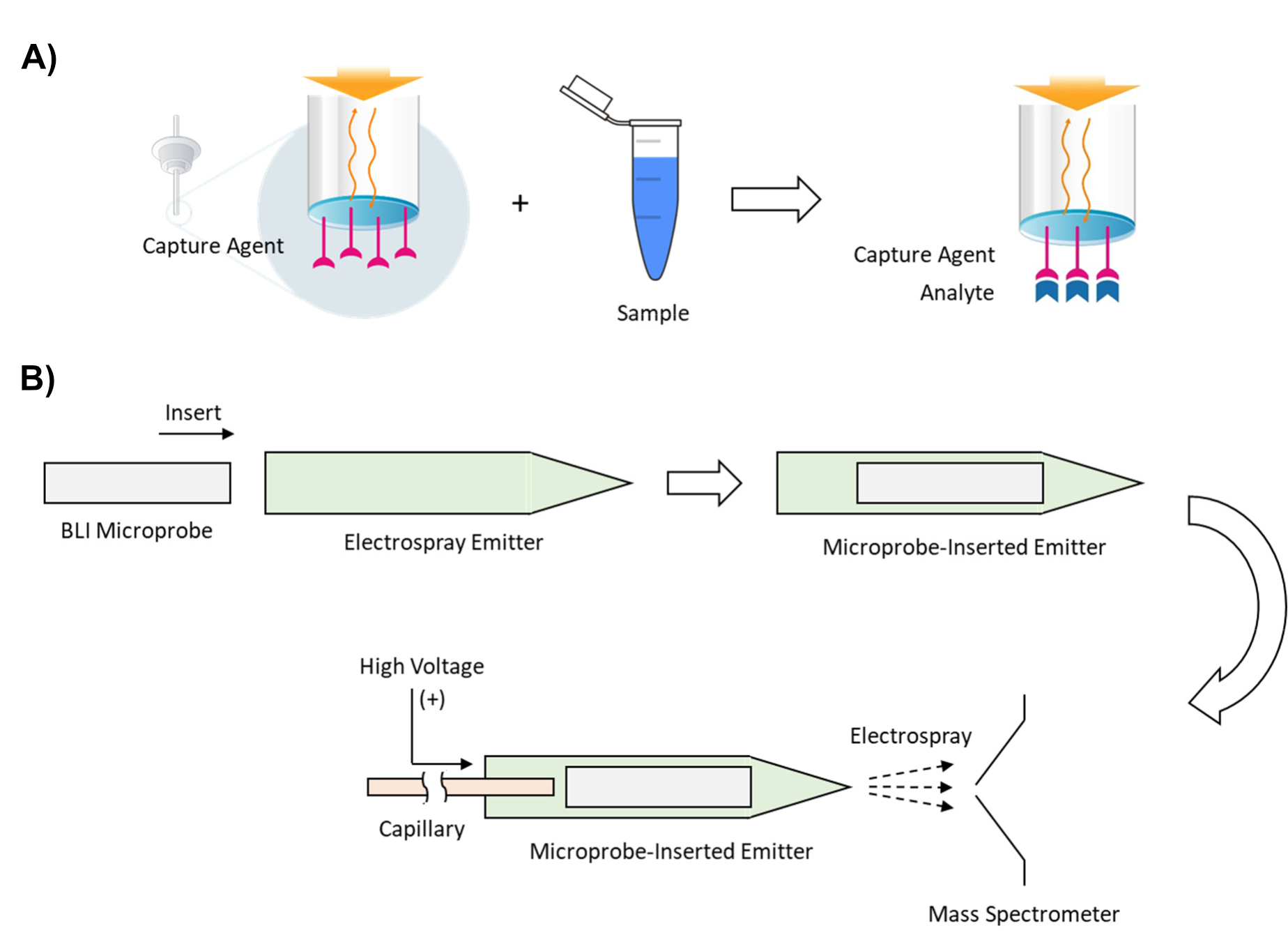
Figure 1. Microprobe-Capture In-Emitter Elution (MPIE) A) Schematic of BLI-based affinity capture on a microprobe B) Schematic of In-Emitter Elution for ESI-MS.
Image credit to the inventors.
Stage of Development Prototype
Applications
- Top-down mass spectrometry
- Antibody epitope mapping
- Affinity selection of protein binders
- Target quantitation
Advantages
- Efficient analyte transfer from the microprobe to the electrospray emitter enables high sensitivity MS analysis
- Affinity capture conditions can be optimized efficiently, without requiring MS analysis
- Estimated amount of captured analyte is acquired for every sample, providing improved quality control
Publications
- Luo R.Y., Yang S. (2022). Microprobe-Capture In-Emitter Elution: An Affinity Capture Technique to Directly Couple a Label-Free Optical Sensing Technology with Mass Spectrometry for Top-Down Protein Analysis. ChemRxiv. 10.26434/chemrxiv-2022-9bjg0-v4
Related Links
Patents
- Published Application: 20230305019
Similar Technologies
-
Wide-field Resonant Electro-optic Imaging Devices and Applications S19-424Wide-field Resonant Electro-optic Imaging Devices and Applications
-
Efficient wide-field nanosecond imaging methods using Pockels cells for low-light applications S18-388Efficient wide-field nanosecond imaging methods using Pockels cells for low-light applications
-
Light sheet fluorescence microscopy using high speed structured and pivoting illumination S16-332Light sheet fluorescence microscopy using high speed structured and pivoting illumination