Docket #: S24-214
Tunable Protein Domains for Enhanced Post-transcriptional Gene Regulation in Human Cells
Stanford scientists in Lacramioara Bintu's lab have developed a high-throughput system to identify regulatory domains in human RNA-binding proteins, presenting a new set of tools that could greatly enhance control over gene regulation at the RNA level for therapeutic and synthetic biology applications.
The field of gene regulation has increasingly identified the crucial role of post-transcriptional regulation via RNA-binding proteins (RBPs). However, until recent advances in RNA targeting and editing technologies, it has been challenging to achieve precise control of gene regulation in human cells at the RNA level. Existing tools often lack compact protein domains that can regulate mRNA fate in a tunable manner. Moreover, most current synthetic RNA tools are constitutive binders, making it difficult to manage gene regulation appropriately.
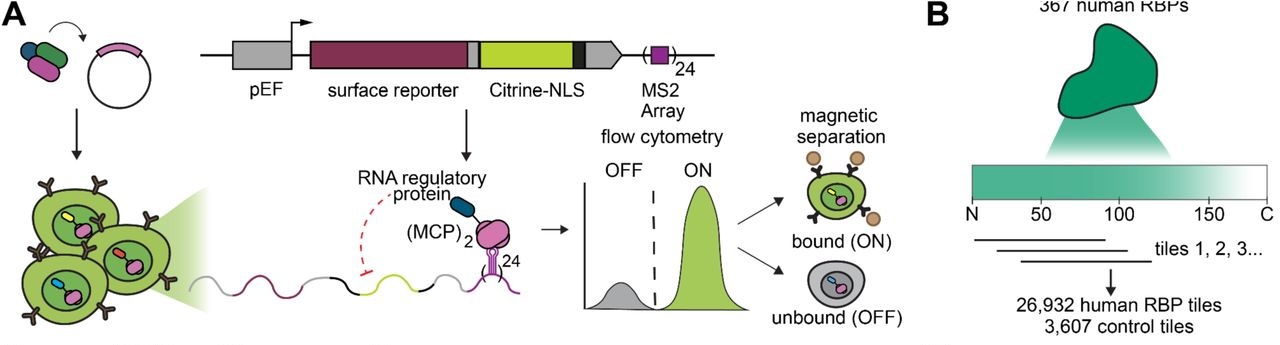
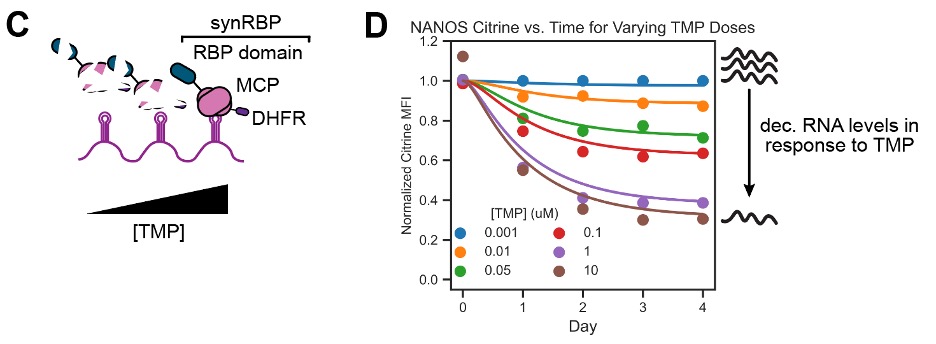
Stanford researchers have created a high-throughput system to identify and catalog RNA regulatory domains in human RNA-binding proteins, a key element in gene expression. This technology facilitates the identification of hundreds of protein domains, which when fused to an RNA-binding domain, can lead to mRNA degradation and prevent translation. The advantage of this system is its tunability, allowing more precise control over mRNA and subsequent protein levels, something that competing technologies and previous methods do not currently offer.
Figure
Stage of Development:
Proof of concept - in vitro
Applications
- Gene and cell therapy: Programmable RNA-based tool to control RNA degradation rates of targeted RNAs
- Synthetic circuits or synthetic clocks: Programmable level of control at the RNA level
- Cholesterol-associated disorders: Careful equilibrium of expressed RNAs
Advantages
- High-throughput
- First in class: The first catalog of RNA-regulating protein tiles from human proteins, advancing synthetic RNA-targeting in human biology and therapeutics
- Tunable: Better dose control compared to existing synthetic RNA tools
- Compact: Discovered tiles are 80 amino acids long, an easily deliverable size for cellular therapeutics.
Publications
- Thurm, A. R., Finkel, Y., Andrews, C., Cai, X. S., Benko, C., & Bintu, L. (2024). High-throughput discovery of regulatory effector domains in human RNA-binding proteins. bioRxiv.
Related Links
Patents
- Published Application: WO2026020078
Similar Technologies
-
A Method for Suppressing Innate Immune Responses to RNA Therapy S22-415A Method for Suppressing Innate Immune Responses to RNA Therapy
-
Myc degradation by long non coding RNA for cancer therapeutics S22-163Myc degradation by long non coding RNA for cancer therapeutics
-
Therapeutic and Immunogenic Protein Production Using Mammalian IRES-like Sequences for Enhanced Circular RNA translation S24-141Therapeutic and Immunogenic Protein Production Using Mammalian IRES-like Sequences for Enhanced Circular RNA translation