Docket #: S17-352
TurboID and miniTurbo- engineered promiscuous biotin ligases for efficient proximity labeling in living cells and organisms
Researchers at Stanford have engineered two promiscuous biotin ligases for non-toxic, efficient proximity labeling (PL) in living cells and organisms. PL is a powerful technique for the proteomic analysis of macromolecular complexes, organelles or protein interaction networks. In PL, a promiscuous labeling enzyme is fused to a protein of interest or targeted to a subcellular region and upon the addition of biotin, the enzyme begins labeling nearby (within a few nanometers) endogenous proteins. Then the biotin labelled proteins are harvested and identified by mass spectrometry. Existing PL techniques are not optimal as they require either toxic regents or long labelling times that prevent the study of dynamic structures or processes. To overcome these limitations the inventors have engineered two mutants of biotin ligase, called TurboID and miniTurbo, which are both more active and efficient than existing variants. TurboID and miniTurbo enable PL with greater signal in shorter time windows under physiologically relevant conditions in live cells. This technology will expand the scope and utility of PL by enabling the study of dynamic processes in vivo and thus can provide insight into proteomic changes to aid general research and therapeutic development.
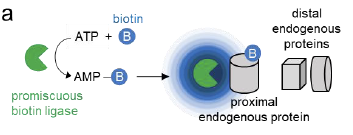
Proximity-dependent biotinylation by promiscuous biotin ligases. Ligases catalyze the formation of biotin-5'-AMP anhydride, which diffuses out of the active site to biotinylate proximal endogenous proteins.
Stage of research
In addition to shortening PL time by 100-fold and increasing PL yield in cell culture, the inventors have shown that TurboID enabled biotin-based PL in new settings including yeast, Drosophila and C. elegans.
Applications
- Proteomic analysis for:
- Research
- Therapeutic development
Advantages
- Efficient- shortens PL time by 100-fold
- Short labeling time-can be used to resolve dynamic proteomic changes during development or in response to drugs or stimuli
- Enhanced activity- proteomic labeling in just 10 mins (rather the 18-24hrs required by other methods)
- Non-toxic- can be used in living cells and organisms
- Allows proteomic mapping under physiologically relevant conditions with high temporal resolution
- Simple labeling protocol
- Can be genetically targeted to any subcellular compartment or fused to any protein of interest
- Allows clinically relevant studies in animal models
Publications
- Branon, T., Bosch, J., Sanchez, A. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887 (2018).
Related Links
Patents
- Published Application: WO2019143529
- Published Application: 20210214708
- Issued: 12,110,522 (USA)
Similar Technologies
-
Method of RNA sample preparation from Ribonuclease-rich matrices S12-276Method of RNA sample preparation from Ribonuclease-rich matrices
-
HCC Cell Line S11-143AHCC Cell Line
-
Non-immunogenic Destabilizing Domains S17-025Non-immunogenic Destabilizing Domains