Docket #: S15-424
RNA fixation and detection in CLARITY-based hydrogel tissue
Researchers in Dr. Karl Deisseroth's lab have patented a versatile and robust RNA fixation and detection method in transparent intact tissues. One of the current challenges in neuroscience is to understand molecular identity, activity level and circuit wiring of individual cells within intact brain networks. Methods to investigate neuronal networks in intact tissues have been developed but are not optimal. For instance, these methods have used antibody staining for molecular analysis. This is costly and has a low success rate. In addition, these approaches may not be compatible with RNA detection and thus a wealth of biological information is left behind. There is a need to develop additional methods to visualize neuronal networks including the RNA in intact tissues. To help meet this need the inventors developed this technology which provides methods for fixation and detection of RNA in transparent intact tissues. These methods allow for cellular-resolution transcriptional profiling of large intact mammalian tissues, with reliable detection of diverse markers for non-coding transcripts, cell identity and activity history.
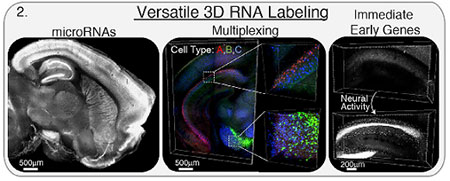
Image courtesy of the Deisseroth Lab
Stage of Development: Other
Deisseroth Lab researchers demonstrated that this technology robustly measures activity-dependent transcriptional signatures, cell-identity markers, and diverse non-coding RNAs in rodent and human tissue. The group uses the technique in ongoing research and actively maintains RNA in CLARITY resources.
Applications
- Tissue and organism research
- Label neuronal structures
- Identify and characterize cell types in intact tissues
- Assess neuronal activation in whole brain context
- Clinical histology
- Analyze spatial changes in RNA expression in disease states
Advantages
- Better preservation of RNA as compared to existing techniques
- Allows simultaneous labeling of RNA products
- Advantageous over protein labeling methods, as it has:
- Scalable molecular phenotyping
- Lower cost
- Availability for all transcripts
- Consistency across populations
- Inclusivity of non-translated mRNA
- Versatile- can use a variety of probe types and amplification methods
- Satisfies unmet need- includes comprehensive set of probes to label mRNAs (including activity-regulated transcripts)
Publications
- Deisseroth, K. A., Sylwestrak, E. L., Rajasethupathy, P., & Wright, M. (2022). U.S. Patent No. 11,254,974. Washington, DC: U.S. Patent and Trademark Office.
- Deisseroth, K. A., Sylwestrak, E. L., Rajasethupathy, P., & Wright, M. (2022). U.S. Patent Application No. 17/570,111.
- Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A., & Deisseroth, K. (2016). Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell, 164(4), 792-804. https://doi.org/10.1016/j.cell.2016.01.038
Related Links
Patents
- Published Application: WO2017139501
- Published Application: 20190119735
- Published Application: 20220389492
- Issued: 11,254,974 (USA)
- Issued: 12,098,418 (USA)
Similar Technologies
-
CLARITY: Transparent Tissue for 3D Imaging of Neuronal Networks and Subcellular Structures S12-036CLARITY: Transparent Tissue for 3D Imaging of Neuronal Networks and Subcellular Structures
-
Light sheet fluorescence microscopy using high speed structured and pivoting illumination S16-332Light sheet fluorescence microscopy using high speed structured and pivoting illumination
-
Light Field Microscope S05-327Light Field Microscope